Mathematical Framework for latentcor
Mingze Huang, Christian L. Müller, Irina Gaynanova
2022-08-31
latentcor_math.RmdLatent Gaussian Copula Model for Mixed Data
latentcor utilizes the powerful semi-parametric latent
Gaussian copula models to estimate latent correlations between mixed
data types (continuous/binary/ternary/truncated or zero-inflated). Below
we review the definitions for each type.
Definition of continuous model (Fan et al. 2017)
A random \(X\in\cal{R}^{p}\) satisfies the Gaussian copula (or nonparanormal) model if there exist monotonically increasing \(f=(f_{j})_{j=1}^{p}\) with \(Z_{j}=f_{j}(X_{j})\) satisfying \(Z\sim N_{p}(0, \Sigma)\), \(\sigma_{jj}=1\); we denote \(X\sim NPN(0, \Sigma, f)\).
X = gen_data(n = 6, types = "con")$X
X
#> [,1]
#> [1,] -0.05894138
#> [2,] 0.61420386
#> [3,] 1.62989498
#> [4,] 1.34911221
#> [5,] 0.39471148
#> [6,] -0.32654291Definition of binary model (Fan et al. 2017)
A random \(X\in\cal{R}^{p}\) satisfies the binary latent Gaussian copula model if there exists \(W\sim NPN(0, \Sigma, f)\) such that \(X_{j}=I(W_{j}>c_{j})\), where \(I(\cdot)\) is the indicator function and \(c_{j}\) are constants.
X = gen_data(n = 6, types = "bin")$X
X
#> [,1]
#> [1,] 0
#> [2,] 0
#> [3,] 1
#> [4,] 1
#> [5,] 1
#> [6,] 0Definition of ternary model (Quan, Booth, and Wells 2018)
A random \(X\in\cal{R}^{p}\) satisfies the ternary latent Gaussian copula model if there exists \(W\sim NPN(0, \Sigma, f)\) such that \(X_{j}=I(W_{j}>c_{j})+I(W_{j}>c'_{j})\), where \(I(\cdot)\) is the indicator function and \(c_{j}<c'_{j}\) are constants.
X = gen_data(n = 6, types = "ter")$X
X
#> [,1]
#> [1,] 0
#> [2,] 2
#> [3,] 1
#> [4,] 1
#> [5,] 1
#> [6,] 0Definition of truncated or zero-inflated model (Yoon, Carroll, and Gaynanova 2020)
A random \(X\in\cal{R}^{p}\) satisfies the truncated latent Gaussian copula model if there exists \(W\sim NPN(0, \Sigma, f)\) such that \(X_{j}=I(W_{j}>c_{j})W_{j}\), where \(I(\cdot)\) is the indicator function and \(c_{j}\) are constants.
X = gen_data(n = 6, types = "tru")$X
X
#> [,1]
#> [1,] 1.1646683
#> [2,] 0.1497381
#> [3,] 0.0000000
#> [4,] 0.0000000
#> [5,] 0.0000000
#> [6,] 0.5315542Mixed latent Gaussian copula model
The mixed latent Gaussian copula model jointly models \(W=(W_{1}, W_{2}, W_{3}, W_{4})\sim NPN(0, \Sigma, f)\) such that \(X_{1j}=W_{1j}\), \(X_{2j}=I(W_{2j}>c_{2j})\), \(X_{3j}=I(W_{3j}>c_{3j})+I(W_{3j}>c'_{3j})\) and \(X_{4j}=I(W_{4j}>c_{4j})W_{4j}\).
set.seed("234820")
X = gen_data(n = 100, types = c("con", "bin", "ter", "tru"))$X
head(X)
#> [,1] [,2] [,3] [,4]
#> [1,] -0.5728663 0 1 0.0000000
#> [2,] -1.5632883 0 0 0.0000000
#> [3,] 0.4600555 1 1 0.2634213
#> [4,] -1.5186510 1 1 0.0000000
#> [5,] -1.5438165 0 1 0.0000000
#> [6,] -0.5656219 0 0 0.0000000Moment-based estimation of \(\Sigma\) based on bridge functions
The estimation of latent correlation matrix \(\Sigma\) is achieved via the bridge function \(F\) which is defined such that \(E(\hat{\tau}_{jk})=F(\sigma_{jk})\), where \(\sigma_{jk}\) is the latent correlation between variables \(j\) and \(k\), and \(\hat{\tau}_{jk}\) is the corresponding sample Kendall’s \(\tau\).
Kendall’s \(\tau\) (\(\tau_{a}\))
Given observed \(\mathbf{x}_{j}, \mathbf{x}_{k}\in\cal{R}^{n}\),
\[ \hat{\tau}_{jk}=\hat{\tau}(\mathbf{x}_{j}, \mathbf{x}_{k})=\frac{2}{n(n-1)}\sum_{1\le i<i'\le n}sign(x_{ij}-x_{i'j})sign(x_{ik}-x_{i'k}), \] where \(n\) is the sample size.
latentcor calculates pairwise Kendall’s \(\widehat \tau\) as part of the estimation
process
estimate = latentcor(X, types = c("con", "bin", "ter", "tru"))
K = estimate$K
K
#> [,1] [,2] [,3] [,4]
#> [1,] 1.0000000 0.2557576 0.2456566 0.3331313
#> [2,] 0.2557576 1.0000000 0.1555556 0.2339394
#> [3,] 0.2456566 0.1555556 1.0000000 0.2183838
#> [4,] 0.3331313 0.2339394 0.2183838 1.0000000Using \(F\) and \(\widehat \tau_{jk}\), a moment-based estimator is \(\hat{\sigma}_{jk}=F^{-1}(\hat{\tau}_{jk})\) with the corresponding \(\hat{\Sigma}\) being consistent for \(\Sigma\) (Fan et al. 2017; Quan, Booth, and Wells 2018; Yoon, Carroll, and Gaynanova 2020).
The explicit form of bridge function \(F\) has been derived for all combinations
of continuous(C)/binary(B)/ternary(N)/truncated(T) variable types, and
we summarize the corresponding references. Each of this combinations is
implemented in latentcor.
| Type | continuous | binary | ternary | zero-inflated (truncated) |
|---|---|---|---|---|
| continuous | Liu, Lafferty, and Wasserman (2009) | - | - | - |
| binary | Fan et al. (2017) | Fan et al. (2017) | - | - |
| ternary | Quan, Booth, and Wells (2018) | Quan, Booth, and Wells (2018) | Quan, Booth, and Wells (2018) | - |
| zero-inflated (truncated) | Yoon, Carroll, and Gaynanova (2020) | Yoon, Carroll, and Gaynanova (2020) | See Appendix | Yoon, Carroll, and Gaynanova (2020) |
Below we provide an explicit form of \(F\) for each combination.
Theorem (explicit form of bridge function) Let \(W_{1}\in\cal{R}^{p_{1}}\), \(W_{2}\in\cal{R}^{p_{2}}\), \(W_{3}\in\cal{R}^{p_{3}}\), \(W_{4}\in\cal{R}^{p_{4}}\) be such that \(W=(W_{1}, W_{2}, W_{3}, W_{4})\sim NPN(0, \Sigma, f)\) with \(p=p_{1}+p_{2}+p_{3}+p_{4}\). Let \(X=(X_{1}, X_{2}, X_{3}, X_{4})\in\cal{R}^{p}\) satisfy \(X_{j}=W_{j}\) for \(j=1,...,p_{1}\), \(X_{j}=I(W_{j}>c_{j})\) for \(j=p_{1}+1, ..., p_{1}+p_{2}\), \(X_{j}=I(W_{j}>c_{j})+I(W_{j}>c'_{j})\) for \(j=p_{1}+p_{2}+1, ..., p_{3}\) and \(X_{j}=I(W_{j}>c_{j})W_{j}\) for \(j=p_{1}+p_{2}+p_{3}+1, ..., p\) with \(\Delta_{j}=f(c_{j})\). The rank-based estimator of \(\Sigma\) based on the observed \(n\) realizations of \(X\) is the matrix \(\mathbf{\hat{R}}\) with \(\hat{r}_{jj}=1\), \(\hat{r}_{jk}=\hat{r}_{kj}=F^{-1}(\hat{\tau}_{jk})\) with block structure
\[ \mathbf{\hat{R}}=\begin{pmatrix} F_{CC}^{-1}(\hat{\tau}) & F_{CB}^{-1}(\hat{\tau}) & F_{CN}^{-1}(\hat{\tau}) & F_{CT}^{-1}(\hat{\tau})\\ F_{BC}^{-1}(\hat{\tau}) & F_{BB}^{-1}(\hat{\tau}) & F_{BN}^{-1}(\hat{\tau}) & F_{BT}^{-1}(\hat{\tau})\\ F_{NC}^{-1}(\hat{\tau}) & F_{NB}^{-1}(\hat{\tau}) & F_{NN}^{-1}(\hat{\tau}) & F_{NT}^{-1}(\hat{\tau})\\ F_{TC}^{-1}(\hat{\tau}) & F_{TB}^{-1}(\hat{\tau}) & F_{TN}^{-1}(\hat{\tau}) & F_{TT}^{-1}(\hat{\tau}) \end{pmatrix} \] \[ F(\cdot)=\begin{cases} CC:\ 2\sin^{-1}(r)/\pi \\ \\ BC: \ 4\Phi_{2}(\Delta_{j},0;r/\sqrt{2})-2\Phi(\Delta_{j}) \\ \\ BB: \ 2\{\Phi_{2}(\Delta_{j},\Delta_{k};r)-\Phi(\Delta_{j})\Phi(\Delta_{k})\} \\ \\ NC: \ 4\Phi_{2}(\Delta_{j}^{2},0;r/\sqrt{2})-2\Phi(\Delta_{j}^{2})+4\Phi_{3}(\Delta_{j}^{1},\Delta_{j}^{2},0;\Sigma_{3a}(r))-2\Phi(\Delta_{j}^{1})\Phi(\Delta_{j}^{2})\\ \\ NB: \ 2\Phi_{2}(\Delta_{j}^{2},\Delta_{k},r)\{1-\Phi(\Delta_{j}^{1})\}-2\Phi(\Delta_{j}^{2})\{\Phi(\Delta_{k})-\Phi_{2}(\Delta_{j}^{1},\Delta_{k},r)\} \\ \\ NN: \ 2\Phi_{2}(\Delta_{j}^{2},\Delta_{k}^{2};r)\Phi_{2}(-\Delta_{j}^{1},-\Delta_{k}^{1};r)-2\{\Phi(\Delta_{j}^{2})-\Phi_{2}(\Delta_{j}^{2},\Delta_{k}^{1};r)\}\{\Phi(\Delta_{k}^{2})-\Phi_{2}(\Delta_{j}^{1},\Delta_{k}^{2};r)\} \\ \\ TC: \ -2\Phi_{2}(-\Delta_{j},0;1/\sqrt{2})+4\Phi_{3}(-\Delta_{j},0,0;\Sigma_{3b}(r)) \\ \\ TB: \ 2\{1-\Phi(\Delta_{j})\}\Phi(\Delta_{k})-2\Phi_{3}(-\Delta_{j},\Delta_{k},0;\Sigma_{3c}(r))-2\Phi_{3}(-\Delta_{j},\Delta_{k},0;\Sigma_{3d}(r)) \\ \\ TN: \ -2\Phi(-\Delta_{k}^{1})\Phi(\Delta_{k}^{2}) + 2\Phi_{3}(-\Delta_{k}^{1},\Delta_{k}^{2},\Delta_{j};\Sigma_{3e}(r))+2\Phi_{4}(-\Delta_{k}^{1},\Delta_{k}^{2},-\Delta_{j},0;\Sigma_{4a}(r))+2\Phi_{4}(-\Delta_{k}^{1},\Delta_{k}^{2},-\Delta_{j},0;\Sigma_{4b}(r)) \\ \\ TT: \ -2\Phi_{4}(-\Delta_{j},-\Delta_{k},0,0;\Sigma_{4c}(r))+2\Phi_{4}(-\Delta_{j},-\Delta_{k},0,0;\Sigma_{4d}(r)) \\ \end{cases} \]
where \(\Delta_{j}=\Phi^{-1}(\pi_{0j})\), \(\Delta_{k}=\Phi^{-1}(\pi_{0k})\), \(\Delta_{j}^{1}=\Phi^{-1}(\pi_{0j})\), \(\Delta_{j}^{2}=\Phi^{-1}(\pi_{0j}+\pi_{1j})\), \(\Delta_{k}^{1}=\Phi^{-1}(\pi_{0k})\), \(\Delta_{k}^{2}=\Phi^{-1}(\pi_{0k}+\pi_{1k})\),
\[ \Sigma_{3a}(r)= \begin{pmatrix} 1 & 0 & \frac{r}{\sqrt{2}} \\ 0 & 1 & -\frac{r}{\sqrt{2}} \\ \frac{r}{\sqrt{2}} & -\frac{r}{\sqrt{2}} & 1 \end{pmatrix}, \;\;\; \Sigma_{3b}(r)= \begin{pmatrix} 1 & \frac{1}{\sqrt{2}} & \frac{r}{\sqrt{2}}\\ \frac{1}{\sqrt{2}} & 1 & r \\ \frac{r}{\sqrt{2}} & r & 1 \end{pmatrix}, \;\;\; \Sigma_{3c}(r)= \begin{pmatrix} 1 & -r & \frac{1}{\sqrt{2}} \\ -r & 1 & -\frac{r}{\sqrt{2}} \\ \frac{1}{\sqrt{2}} & -\frac{r}{\sqrt{2}} & 1 \end{pmatrix}, \]
\[ \Sigma_{3d}(r)= \begin{pmatrix} 1 & 0 & -\frac{1}{\sqrt{2}} \\ 0 & 1 & -\frac{r}{\sqrt{2}} \\ -\frac{1}{\sqrt{2}} & -\frac{r}{\sqrt{2}} & 1 \end{pmatrix}, \;\;\; \Sigma_{3e}(r)= \begin{pmatrix} 1 & 0 & 0 \\ 0 & 1 & r \\ 0 & r & 1 \end{pmatrix}, \;\;\; \Sigma_{4a}(r)= \begin{pmatrix} 1 & 0 & 0 & \frac{r}{\sqrt{2}} \\ 0 & 1 & -r & \frac{r}{\sqrt{2}} \\ 0 & -r & 1 & -\frac{1}{\sqrt{2}} \\ \frac{r}{\sqrt{2}} & \frac{r}{\sqrt{2}} & -\frac{1}{\sqrt{2}} & 1 \end{pmatrix}, \]
\[ \Sigma_{4b}(r)= \begin{pmatrix} 1 & 0 & r & \frac{r}{\sqrt{2}} \\ 0 & 1 & 0 & \frac{r}{\sqrt{2}} \\ r & 0 & 1 & \frac{1}{\sqrt{2}} \\ \frac{r}{\sqrt{2}} & \frac{r}{\sqrt{2}} & \frac{1}{\sqrt{2}} & 1 \end{pmatrix}, \;\;\; \Sigma_{4c}(r)= \begin{pmatrix} 1 & 0 & \frac{1}{\sqrt{2}} & -\frac{r}{\sqrt{2}} \\ 0 & 1 & -\frac{r}{\sqrt{2}} & \frac{1}{\sqrt{2}} \\ \frac{1}{\sqrt{2}} & -\frac{r}{\sqrt{2}} & 1 & -r \\ -\frac{r}{\sqrt{2}} & \frac{1}{\sqrt{2}} & -r & 1 \end{pmatrix}\;\;\text{and}\;\; \Sigma_{4d}(r)= \begin{pmatrix} 1 & r & \frac{1}{\sqrt{2}} & \frac{r}{\sqrt{2}} \\ r & 1 & \frac{r}{\sqrt{2}} & \frac{1}{\sqrt{2}} \\ \frac{1}{\sqrt{2}} & \frac{r}{\sqrt{2}} & 1 & r \\ \frac{r}{\sqrt{2}} & \frac{1}{\sqrt{2}} & r & 1 \end{pmatrix}. \]
Estimation methods
Given the form of bridge function \(F\), obtaining a moment-based estimation
\(\widehat \sigma_{jk}\) requires
inversion of \(F\).
latentcor implements two methods for calculation of the
inversion:
-
method = "original"Subsection describing original method and relevant parametertol -
method = "approx"Subsection describing approximation method and relevant parameterratio
Both methods calculate inverse bridge function applied to each
element of sample Kendall’s \(\tau\)
matrix. Because the calculation is performed point-wise (separately for
each pair of variables), the resulting point-wise estimator of
correlation matrix may not be positive semi-definite.
latentcor performs projection of the pointwise-estimator to
the space of positive semi-definite matrices, and allows for shrinkage
towards identity matrix using the parameter nu (see Subsection describing adjustment of point-wise
estimator and relevant parameter nu).
Original method (method = "original")
Original estimation approach relies on numerical inversion of \(F\) based on solving uni-root optimization
problem. Given the calculated \(\widehat
\tau_{jk}\) (sample Kendall’s \(\tau\) between variables \(j\) and \(k\)), the estimate of latent correlation
\(\widehat \sigma_{jk}\) is obtained by
calling optimize function to solve the following
optimization problem: \[
\widehat r_{jk} = \arg\min_{r} \{F(r) - \widehat \tau_{jk}\}^2.
\] The parameter tol controls the desired accuracy
of the minimizer and is passed to optimize, with the
default precision of 1e-8:
Algorithm for Original method
Input: \(F(r)=F(r, \mathbf{\Delta})\) - bridge function based on the type of variables \(j\), \(k\)
- Step 1. Calculate \(\hat{\tau}_{jk}\) using (1).
estimate$K
#> [,1] [,2] [,3] [,4]
#> [1,] 1.0000000 0.2557576 0.2456566 0.3331313
#> [2,] 0.2557576 1.0000000 0.1555556 0.2339394
#> [3,] 0.2456566 0.1555556 1.0000000 0.2183838
#> [4,] 0.3331313 0.2339394 0.2183838 1.0000000- Step 2. For binary/truncated variable \(j\), set \(\hat{\mathbf{\Delta}}_{j}=\hat{\Delta}_{j}=\Phi^{-1}(\pi_{0j})\) with \(\pi_{0j}=\sum_{i=1}^{n}\frac{I(x_{ij}=0)}{n}\). For ternary variable \(j\), set \(\hat{\mathbf{\Delta}}_{j}=(\hat{\Delta}_{j}^{1}, \hat{\Delta}_{j}^{2})\) where \(\hat{\Delta}_{j}^{1}=\Phi^{-1}(\pi_{0j})\) and \(\hat{\Delta}_{j}^{2}=\Phi^{-1}(\pi_{0j}+\pi_{1j})\) with \(\pi_{0j}=\sum_{i=1}^{n}\frac{I(x_{ij}=0)}{n}\) and \(\pi_{1j}=\sum_{i=1}^{n}\frac{I(x_{ij}=1)}{n}\).
estimate$zratios
#> [[1]]
#> [1] NA
#>
#> [[2]]
#> [1] 0.5
#>
#> [[3]]
#> [1] 0.3 0.8
#>
#> [[4]]
#> [1] 0.5- Compute \(F^{-1}(\hat{\tau}_{jk})\)
as \(\hat{r}_{jk}=argmin\{F(r)-\hat{\tau}_{jk}\}^{2}\)
solved via
optimizefunction in R with accuracytol.
estimate$Rpointwise
#> [,1] [,2] [,3] [,4]
#> [1,] 1.0000000 0.5529903 0.4480984 0.5826171
#> [2,] 0.5529903 1.0000000 0.4050223 0.5821513
#> [3,] 0.4480984 0.4050223 1.0000000 0.4653875
#> [4,] 0.5826171 0.5821513 0.4653875 1.0000000Approximation method (method = "approx")
A faster approximation method is based on multi-linear interpolation of pre-computed inverse bridge function on a fixed grid of points (Yoon, Müller, and Gaynanova 2021). This is possible as the inverse bridge function is an analytic function of at most 5 parameters:
- Kendall’s \(\tau\)
- Proportion of zeros in the 1st variable
- (Possibly) proportion of zeros and ones in the 1st variable
- (Possibly) proportion of zeros in the 2nd variable
- (Possibly) proportion of zeros and ones in the 2nd variable
In short, d-dimensional multi-linear interpolation uses a weighted
average of \(2^{d}\) neighbors to
approximate the function values at the points within the d-dimensional
cube of the neighbors, and to perform interpolation,
latentcor takes advantage of the R package
chebpol (Gaure 2019). This
approximation method has been first described in (Yoon, Müller, and Gaynanova 2021) for
continuous/binary/truncated cases. In latentcor, we
additionally implement ternary case, and optimize the choice of grid as
well as interpolation boundary for faster computations with smaller
memory footprint.
#estimate = latentcor(X, types = c("con", "bin", "ter", "tru"), method = "approx")Algorithm for Approximation method
Input: Let \(\check{g}=h(g)\), pre-computed values \(F^{-1}(h^{-1}(\check{g}))\) on a fixed grid \(\check{g}\in\check{\cal{G}}\) based on the type of variables \(j\) and \(k\). For binary/continuous case, \(\check{g}=(\check{\tau}_{jk}, \check{\Delta}_{j})\); for binary/binary case, \(\check{g}=(\check{\tau}_{jk}, \check{\Delta}_{j}, \check{\Delta}_{k})\); for truncated/continuous case, \(\check{g}=(\check{\tau}_{jk}, \check{\Delta}_{j})\); for truncated/truncated case, \(\check{g}=(\check{\tau}_{jk}, \check{\Delta}_{j}, \check{\Delta}_{k})\); for ternary/continuous case, \(\check{g}=(\check{\tau}_{jk}, \check{\Delta}_{j}^{1}, \check{\Delta}_{j}^{2})\); for ternary/binary case, \(\check{g}=(\check{\tau}_{jk}, \check{\Delta}_{j}^{1}, \check{\Delta}_{j}^{2}, \check{\Delta}_{k})\); for ternary/truncated case, \(\check{g}=(\check{\tau}_{jk}, \check{\Delta}_{j}^{1}, \check{\Delta}_{j}^{2}, \check{\Delta}_{k})\); for ternay/ternary case, \(\check{g}=(\check{\tau}_{jk}, \check{\Delta}_{j}^{1}, \check{\Delta}_{j}^{2}, \check{\Delta}_{k}^{1}, \check{\Delta}_{k}^{2})\).
Step 1 and Step 2 same as Original method.
Step 3. If \(|\hat{\tau}_{jk}|\le \mbox{ratio}\times \bar{\tau}_{jk}(\cdot)\), apply interpolation; otherwise apply Original method.
To avoid interpolation in areas with high approximation errors close
to the boundary, we use hybrid scheme in Step 3. The parameter
ratio controls the size of the region where the
interpolation is performed (ratio = 0 means no
interpolation, ratio = 1 means interpolation is always
performed). For the derivation of approximate bound for BC, BB, TC, TB,
TT cases see Yoon, Müller, and Gaynanova
(2021). The derivation of approximate bound for NC, NB, NN, NT
case is in the Appendix.
\[ \bar{\tau}_{jk}(\cdot)= \begin{cases} 2\pi_{0j}(1-\pi_{0j}) & for \; BC \; case\\ 2\min(\pi_{0j},\pi_{0k})\{1-\max(\pi_{0j}, \pi_{0k})\} & for \; BB \; case\\ 2\{\pi_{0j}(1-\pi_{0j})+\pi_{1j}(1-\pi_{0j}-\pi_{1j})\} & for \; NC \; case\\ 2\min(\pi_{0j}(1-\pi_{0j})+\pi_{1j}(1-\pi_{0j}-\pi_{1j}),\pi_{0k}(1-\pi_{0k})) & for \; NB \; case\\ 2\min(\pi_{0j}(1-\pi_{0j})+\pi_{1j}(1-\pi_{0j}-\pi_{1j}), \\ \;\;\;\;\;\;\;\;\;\;\pi_{0k}(1-\pi_{0k})+\pi_{1k}(1-\pi_{0k}-\pi_{1k})) & for \; NN \; case\\ 1-(\pi_{0j})^{2} & for \; TC \; case\\ 2\max(\pi_{0k},1-\pi_{0k})\{1-\max(\pi_{0k},1-\pi_{0k},\pi_{0j})\} & for \; TB \; case\\ 1-\{\max(\pi_{0j},\pi_{0k},\pi_{1k},1-\pi_{0k}-\pi_{1k})\}^{2} & for \; TN \; case\\ 1-\{\max(\pi_{0j},\pi_{0k})\}^{2} & for \; TT \; case\\ \end{cases} \]
By default, latentcor uses ratio = 0.9 as
this value was recommended in Yoon, Müller, and
Gaynanova (2021) having a good balance of accuracy and
computational speed. This value, however, can be modified by the
user.
#latentcor(X, types = c("con", "bin", "ter", "tru"), method = "approx", ratio = 0.99)$R
#latentcor(X, types = c("con", "bin", "ter", "tru"), method = "approx", ratio = 0.4)$R
latentcor(X, types = c("con", "bin", "ter", "tru"), method = "original")$R
#> [,1] [,2] [,3] [,4]
#> [1,] 1.0000000 0.5524373 0.4476503 0.5820345
#> [2,] 0.5524373 1.0000000 0.4046173 0.5815691
#> [3,] 0.4476503 0.4046173 1.0000000 0.4649222
#> [4,] 0.5820345 0.5815691 0.4649222 1.0000000The lower is the ratio, the closer is the approximation
method to original method (with ratio = 0 being equivalent
to method = "original"), but also the higher is the cost of
computations.
library(microbenchmark)
#microbenchmark(latentcor(X, types = c("con", "bin", "ter", "tru"), method = "approx", ratio = 0.99)$R)
#microbenchmark(latentcor(X, types = c("con", "bin", "ter", "tru"), method = "approx", ratio = 0.4)$R)
microbenchmark(latentcor(X, types = c("con", "bin", "ter", "tru"), method = "original")$R)
#> Unit: milliseconds
#> expr
#> latentcor(X, types = c("con", "bin", "ter", "tru"), method = "original")$R
#> min lq mean median uq max neval
#> 29.6928 30.16525 30.8096 30.53385 31.0322 35.1031 100Rescaled Grid for Interpolation
Since \(|\hat{\tau}|\le \bar{\tau}\), the grid does not need to cover the whole domain \(\tau\in[-1, 1]\). To optimize memory associated with storing the grid, we rescale \(\tau\) as follows: \(\check{\tau}_{jk}=\tau_{jk}/\bar{\tau}_{jk}\in[-1, 1]\), where \(\bar{\tau}_{jk}\) is as defined above.
In addition, for ternary variable \(j\), it always holds that \(\Delta_{j}^{2}>\Delta_{j}^{1}\) since \(\Delta_{j}^{1}=\Phi^{-1}(\pi_{0j})\) and \(\Delta_{j}^{2}=\Phi^{-1}(\pi_{0j}+\pi_{1j})\). Thus, the grid should not cover the the area corresponding to \(\Delta_{j}^{2}\le\Delta_{j}^{1}\). We thus rescale as follows: \(\check{\Delta}_{j}^{1}=\Delta_{j}^{1}/\Delta_{j}^{2}\in[0, 1]\); \(\check{\Delta}_{j}^{2}=\Delta_{j}^{2}\in[0, 1]\).
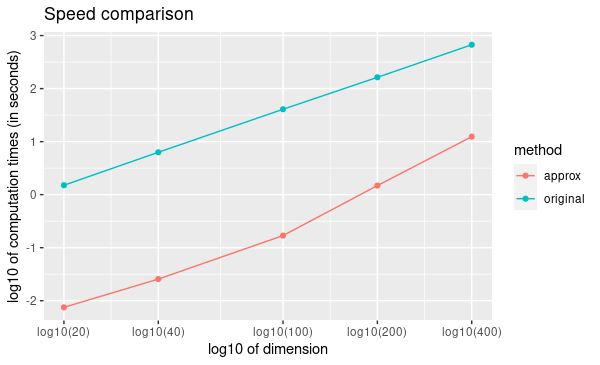
Speed Comparison
To illustrate the speed improvement by
method = "approx", we plot the run time scaling behavior of
method = "approx" and method = "original"
(setting types for gen_data by replicating
c("con", "bin", "ter", "tru") multiple times) with
increasing dimensions \(p = [20, 40, 100, 200,
400]\) at sample size \(n =
100\) using simulation data. Figure below summarizes the observed
scaling in a log-log plot. For both methods we observe the expected
\(O(p^2)\) scaling behavior with
dimension p, i.e., a linear scaling in the log-log plot. However,
method = "approx" is at least one order of magnitude faster
than method = "original" independent of the dimension of
the problem.
Adjustment of pointwise-estimator for positive-definiteness
Since the estimation is performed point-wise, the resulting matrix of estimated latent correlations is not guaranteed to be positive semi-definite. For example, this could be expected when the sample size is small (and so the estimation error for each pairwise correlation is larger)
set.seed("234820")
X = gen_data(n = 6, types = c("con", "bin", "ter", "tru"))$X
X
#> [,1] [,2] [,3] [,4]
#> [1,] -0.5182800 0 1 0.1021738
#> [2,] -1.3017092 0 0 0.0000000
#> [3,] 0.3145191 1 2 0.4213514
#> [4,] -0.6093291 0 1 1.2771610
#> [5,] -1.3175490 1 0 0.0000000
#> [6,] -0.7807245 1 1 0.0000000
out = latentcor(X, types = c("con", "bin", "ter", "tru"))
out$Rpointwise
#> [,1] [,2] [,3] [,4]
#> [1,] 1.0000000 -0.1477240 0.9990000 0.8548518
#> [2,] -0.1477240 1.0000000 0.3523666 -0.5030324
#> [3,] 0.9990000 0.3523666 1.0000000 0.9114307
#> [4,] 0.8548518 -0.5030324 0.9114307 1.0000000
eigen(out$Rpointwise)$values
#> [1] 2.85954424 1.29130852 0.09944544 -0.25029820latentcor automatically corrects the pointwise estimator
to be positive definite by making two adjustments. First, if
Rpointwise has smallest eigenvalue less than zero, the
latentcor projects this matrix to the nearest positive
semi-definite matrix. The user is notified of this adjustment through
the message (supressed in previous code chunk), e.g.
Second, latentcor shrinks the adjusted matrix of
correlations towards identity matrix using the parameter \(\nu\) with default value of 0.001
(nu = 0.001), so that the resulting R is
strictly positive definite with the minimal eigenvalue being greater or
equal to \(\nu\). That is \[
R = (1 - \nu) \widetilde R + \nu I,
\] where \(\widetilde R\) is the
nearest positive semi-definite matrix to Rpointwise.
out = latentcor(X, types = c("con", "bin", "ter", "tru"), nu = 0.001)
out$Rpointwise
#> [,1] [,2] [,3] [,4]
#> [1,] 1.0000000 -0.1477240 0.9990000 0.8548518
#> [2,] -0.1477240 1.0000000 0.3523666 -0.5030324
#> [3,] 0.9990000 0.3523666 1.0000000 0.9114307
#> [4,] 0.8548518 -0.5030324 0.9114307 1.0000000
out$R
#> [,1] [,2] [,3] [,4]
#> [1,] 1.0000000 -0.1053533 0.9232992 0.9048072
#> [2,] -0.1053533 1.0000000 0.2372115 -0.4244433
#> [3,] 0.9232992 0.2372115 1.0000000 0.7723678
#> [4,] 0.9048072 -0.4244433 0.7723678 1.0000000As a result, R and Rpointwise could be
quite different when sample size \(n\)
is small. When \(n\) is large and \(p\) is moderate, the difference is
typically driven by parameter nu.
set.seed("234820")
X = gen_data(n = 100, types = c("con", "bin", "ter", "tru"))$X
out = latentcor(X, types = c("con", "bin", "ter", "tru"), nu = 0.001)
out$Rpointwise
#> [,1] [,2] [,3] [,4]
#> [1,] 1.0000000 0.5528213 0.4476819 0.5823120
#> [2,] 0.5528213 1.0000000 0.4058967 0.5808889
#> [3,] 0.4476819 0.4058967 1.0000000 0.4567771
#> [4,] 0.5823120 0.5808889 0.4567771 1.0000000
out$R
#> [,1] [,2] [,3] [,4]
#> [1,] 1.0000000 0.5522684 0.4472342 0.5817297
#> [2,] 0.5522684 1.0000000 0.4054908 0.5803080
#> [3,] 0.4472342 0.4054908 1.0000000 0.4563203
#> [4,] 0.5817297 0.5803080 0.4563203 1.0000000Appendix
Derivation of bridge function \(F\) for ternary/truncated case
Without loss of generality, let \(j=1\) and \(k=2\). By the definition of Kendall’s \(\tau\), \[ \tau_{12}=E(\hat{\tau}_{12})=E[\frac{2}{n(n-1)}\sum_{1\leq i\leq i' \leq n} sign\{(X_{i1}-X_{i'1})(X_{i2}-X_{i'2})\}]. \] Since \(X_{1}\) is ternary, \[\begin{align} &sign(X_{1}-X_{1}') \nonumber\\ =&[I(U_{1}>C_{11},U_{1}'\leq C_{11})+I(U_{1}>C_{12},U_{1}'\leq C_{12})-I(U_{1}>C_{12},U_{1}'\leq C_{11})] \nonumber\\ &-[I(U_{1}\leq C_{11}, U_{1}'>C_{11})+I(U_{1}\leq C_{12}, U_{1}'>C_{12})-I(U_{1}\leq C_{11}, U_{1}'>C_{12})] \nonumber\\ =&[I(U_{1}>C_{11})-I(U_{1}>C_{11},U_{1}'>C_{11})+I(U_{1}>C_{12})-I(U_{1}>C_{12},U_{1}'>C_{12}) \nonumber\\ &-I(U_{1}>C_{12})+I(U_{1}>C_{12},U_{1}'>C_{11})] \nonumber\\ &-[I(U_{1}'>C_{11})-I(U_{1}>C_{11},U_{1}'>C_{11})+I(U_{1}'>C_{12})-I(U_{1}>C_{12},U_{1}'>C_{12}) \nonumber\\ &-I(U_{1}'>C_{12})+I(U_{1}>C_{11},U_{1}'>C_{12})] \nonumber\\ =&I(U_{1}>C_{11})+I(U_{1}>C_{12},U_{1}'>C_{11})-I(U_{1}'>C_{11})-I(U_{1}>C_{11},U_{1}'>C_{12}) \nonumber\\ =&I(U_{1}>C_{11},U_{1}'\leq C_{12})-I(U_{1}'>C_{11},U_{1}\leq C_{12}). \end{align}\] Since \(X_{2}\) is truncated, \(C_{1}>0\) and \[\begin{align} sign(X_{2}-X_{2}')=&-I(X_{2}=0,X_{2}'>0)+I(X_{2}>0,X_{2}'=0) \nonumber\\ &+I(X_{2}>0,X_{2}'>0)sign(X_{2}-X_{2}') \nonumber\\ =&-I(X_{2}=0)+I(X_{2}'=0)+I(X_{2}>0,X_{2}'>0)sign(X_{2}-X_{2}'). \end{align}\] Since \(f\) is monotonically increasing, \(sign(X_{2}-X_{2}')=sign(Z_{2}-Z_{2}')\), \[\begin{align} \tau_{12}=&E[I(U_{1}>C_{11},U_{1}'\leq C_{12}) sign(X_{2}-X_{2}')] \nonumber\\ &-E[I(U_{1}'>C_{11},U_{1}\leq C_{12}) sign(X_{2}-X_{2}')] \nonumber\\ =&-E[I(U_{1}>C_{11},U_{1}'\leq C_{12}) I(X_{2}=0)] \nonumber\\ &+E[I(U_{1}>C_{11},U_{1}'\leq C_{12}) I(X_{2}'=0)] \nonumber\\ &+E[I(U_{1}>C_{11},U_{1}'\leq C_{12})I(X_{2}>0,X_{2}'>0)sign(Z_{2}-Z_{2}')] \nonumber\\ &+E[I(U_{1}'>C_{11},U_{1}\leq C_{12}) I(X_{2}=0)] \nonumber\\ &-E[I(U_{1}'>C_{11},U_{1}\leq C_{12}) I(X_{2}'=0)] \nonumber\\ &-E[I(U_{1}'>C_{11},U_{1}\leq C_{12})I(X_{2}>0,X_{2}'>0)sign(Z_{2}-Z_{2}')] \nonumber\\ =&-2E[I(U_{1}>C_{11},U_{1}'\leq C_{12}) I(X_{2}=0)] \nonumber\\ &+2E[I(U_{1}>C_{11},U_{1}'\leq C_{12}) I(X_{2}'=0)] \nonumber\\ &+E[I(U_{1}>C_{11},U_{1}'\leq C_{12})I(X_{2}>0,X_{2}'>0)sign(Z_{2}-Z_{2}')] \nonumber\\ &-E[I(U_{1}'>C_{11},U_{1}\leq C_{12})I(X_{2}>0,X_{2}'>0)sign(Z_{2}-Z_{2}')]. \end{align}\] From the definition of \(U\), let \(Z_{j}=f_{j}(U_{j})\) and \(\Delta_{j}=f_{j}(C_{j})\) for \(j=1,2\). Using \(sign(x)=2I(x>0)-1\), we obtain \[\begin{align} \tau_{12}=&-2E[I(Z_{1}>\Delta_{11},Z_{1}'\leq \Delta_{12},Z_{2}\leq \Delta_{2})]+2E[I(Z_{1}>\Delta_{11},Z_{1}'\leq \Delta_{12},Z_{2}'\leq \Delta_{2})] \nonumber\\ &+2E[I(Z_{1}>\Delta_{11},Z_{1}'\leq \Delta_{12})I(Z_{2}>\Delta_{2},Z_{2}'>\Delta_{2},Z_{2}-Z_{2}'>0)] \nonumber\\ &-2E[I(Z_{1}'>\Delta_{11},Z_{1}\leq \Delta_{12})I(Z_{2}>\Delta_{2},Z_{2}'>\Delta_{2},Z_{2}-Z_{2}'>0)] \nonumber\\ =&-2E[I(Z_{1}>\Delta_{11},Z_{1}'\leq \Delta_{12}, Z_{2}\leq \Delta_{2})]+2E[I(Z_{1}>\Delta_{11},Z_{1}'\leq \Delta_{12}, Z_{2}'\leq \Delta_{2})] \nonumber\\ &+2E[I(Z_{1}>\Delta_{11},Z_{1}'\leq\Delta_{12},Z_{2}'>\Delta_{2},Z_{2}>Z_{2}')] \nonumber\\ &-2E[I(Z_{1}'>\Delta_{11},Z_{1}\leq\Delta_{12},Z_{2}'>\Delta_{2},Z_{2}>Z_{2}')]. \end{align}\] Since \(\{\frac{Z_{2}'-Z_{2}}{\sqrt{2}}, -Z{1}\}\), \(\{\frac{Z_{2}'-Z_{2}}{\sqrt{2}}, Z{1}'\}\) and \(\{\frac{Z_{2}'-Z_{2}}{\sqrt{2}}, -Z{2}'\}\) are standard bivariate normally distributed variables with correlation \(-\frac{1}{\sqrt{2}}\), \(r/\sqrt{2}\) and \(-\frac{r}{\sqrt{2}}\), respectively, by the definition of \(\Phi_3(\cdot,\cdot, \cdot;\cdot)\) and \(\Phi_4(\cdot,\cdot, \cdot,\cdot;\cdot)\) we have \[\begin{align} F_{NT}(r;\Delta_{j}^{1},\Delta_{j}^{2},\Delta_{k})= & -2\Phi_{3}\left\{-\Delta_{j}^{1},\Delta_{j}^{2},\Delta_{k};\begin{pmatrix} 1 & 0 & -r \\ 0 & 1 & 0 \\ -r & 0 & 1 \end{pmatrix} \right\} \nonumber\\ &+2\Phi_{3}\left\{-\Delta_{j}^{1},\Delta_{j}^{2},\Delta_{k};\begin{pmatrix} 1 & 0 & 0 \\ 0 & 1 & r \\ 0 & r & 1 \end{pmatrix}\right\}\nonumber \\ & +2\Phi_{4}\left\{-\Delta_{j}^{1},\Delta_{j}^{2},-\Delta_{k},0;\begin{pmatrix} 1 & 0 & 0 & \frac{r}{\sqrt{2}} \\ 0 & 1 & -r & \frac{r}{\sqrt{2}} \\ 0 & -r & 1 & -\frac{1}{\sqrt{2}} \\ \frac{r}{\sqrt{2}} & \frac{r}{\sqrt{2}} & -\frac{1}{\sqrt{2}} & 1 \end{pmatrix}\right\} \nonumber\\ &-2\Phi_{4}\left\{-\Delta_{j}^{1},\Delta_{j}^{2},-\Delta_{k},0;\begin{pmatrix} 1 & 0 & r & -\frac{r}{\sqrt{2}} \\ 0 & 1 & 0 & -\frac{r}{\sqrt{2}} \\ r & 0 & 1 & -\frac{1}{\sqrt{2}} \\ -\frac{r}{\sqrt{2}} & -\frac{r}{\sqrt{2}} & -\frac{1}{\sqrt{2}} & 1 \end{pmatrix}\right\}. \end{align}\] Using the facts that \[\begin{align} &\Phi_{4}\left\{-\Delta_{j}^{1},\Delta_{j}^{2},-\Delta_{k},0;\begin{pmatrix} 1 & 0 & r & -\frac{r}{\sqrt{2}} \\ 0 & 1 & 0 & -\frac{r}{\sqrt{2}} \\ r & 0 & 1 & -\frac{1}{\sqrt{2}} \\ -\frac{r}{\sqrt{2}} & -\frac{r}{\sqrt{2}} & -\frac{1}{\sqrt{2}} & 1 \end{pmatrix}\right\} \nonumber\\ &+\Phi_{4}\left\{-\Delta_{j}^{1},\Delta_{j}^{2},-\Delta_{k},0;\begin{pmatrix} 1 & 0 & r & \frac{r}{\sqrt{2}} \\ 0 & 1 & 0 & \frac{r}{\sqrt{2}} \\ r & 0 & 1 & \frac{1}{\sqrt{2}} \\ \frac{r}{\sqrt{2}} & \frac{r}{\sqrt{2}} & \frac{1}{\sqrt{2}} & 1 \end{pmatrix}\right\} \nonumber\\ =&\Phi_{3}\left\{-\Delta_{j}^{1},\Delta_{j}^{2},-\Delta_{k};\begin{pmatrix} 1 & 0 & 0 \\ 0 & 1 & r \\ 0 & r & 1 \end{pmatrix}\right\} \end{align}\] and \[\begin{align} &\Phi_{3}\left\{-\Delta_{j}^{1},\Delta_{j}^{2},-\Delta_{k};\begin{pmatrix} 1 & 0 & 0 \\ 0 & 1 & r \\ 0 & r & 1 \end{pmatrix}\right\}+\Phi_{3}\left\{-\Delta_{j}^{1},\Delta_{j}^{2},\Delta_{k};\begin{pmatrix} 1 & 0 & -r \\ 0 & 1 & 0 \\ -r & 0 & 1 \end{pmatrix} \right\} \nonumber\\ =&\Phi_{2}(-\Delta_{j}^{1},\Delta_{j}^{2};0) =\Phi(-\Delta_{j}^{1})\Phi(\Delta_{j}^{2}). \end{align}\] So that, \[\begin{align} F_{NT}(r;\Delta_{j}^{1},\Delta_{j}^{2},\Delta_{k})= & -2\Phi(-\Delta_{j}^{1})\Phi(\Delta_{j}^{2}) \nonumber\\ &+2\Phi_{3}\left\{-\Delta_{j}^{1},\Delta_{j}^{2},\Delta_{k};\begin{pmatrix} 1 & 0 & 0 \\ 0 & 1 & r \\ 0 & r & 1 \end{pmatrix}\right\}\nonumber \\ & +2\Phi_{4}\left\{-\Delta_{j}^{1},\Delta_{j}^{2},-\Delta_{k},0;\begin{pmatrix} 1 & 0 & 0 & \frac{r}{\sqrt{2}} \\ 0 & 1 & -r & \frac{r}{\sqrt{2}} \\ 0 & -r & 1 & -\frac{1}{\sqrt{2}} \\ \frac{r}{\sqrt{2}} & \frac{r}{\sqrt{2}} & -\frac{1}{\sqrt{2}} & 1 \end{pmatrix}\right\} \nonumber\\ &+2\Phi_{4}\left\{-\Delta_{j}^{1},\Delta_{j}^{2},-\Delta_{k},0;\begin{pmatrix} 1 & 0 & r & \frac{r}{\sqrt{2}} \\ 0 & 1 & 0 & \frac{r}{\sqrt{2}} \\ r & 0 & 1 & \frac{1}{\sqrt{2}} \\ \frac{r}{\sqrt{2}} & \frac{r}{\sqrt{2}} & \frac{1}{\sqrt{2}} & 1 \end{pmatrix}\right\}. \end{align}\]
It is easy to get the bridge function for truncated/ternary case by switching \(j\) and \(k\).
Derivation of approximate bound for the ternary/continuous case
Let \(n_{0x}=\sum_{i=1}^{n_x}I(x_{i}=0)\), \(n_{2x}=\sum_{i=1}^{n_x}I(x_{i}=2)\), \(\pi_{0x}=\frac{n_{0x}}{n_{x}}\) and \(\pi_{2x}=\frac{n_{2x}}{n_{x}}\), then \[\begin{align} |\tau(\mathbf{x})|\leq & \frac{n_{0x}(n-n_{0x})+n_{2x}(n-n_{0x}-n_{2x})}{\begin{pmatrix} n \\ 2 \end{pmatrix}} \nonumber\\ = & 2\{\frac{n_{0x}}{n-1}-(\frac{n_{0x}}{n})(\frac{n_{0x}}{n-1})+\frac{n_{2x}}{n-1}-(\frac{n_{2x}}{n})(\frac{n_{0x}}{n-1})-(\frac{n_{2x}}{n})(\frac{n_{2x}}{n-1})\} \nonumber\\ \approx & 2\{\frac{n_{0x}}{n}-(\frac{n_{0x}}{n})^2+\frac{n_{2x}}{n}-(\frac{n_{2x}}{n})(\frac{n_{0x}}{n})-(\frac{n_{2x}}{n})^2\} \nonumber\\ = & 2\{\pi_{0x}(1-\pi_{0x})+\pi_{2x}(1-\pi_{0x}-\pi_{2x})\} \end{align}\]
For ternary/binary and ternary/ternary cases, we combine the two individual bounds.
Derivation of approximate bound for the ternary/truncated case
Let \(\mathbf{x}\in\mathcal{R}^{n}\) and \(\mathbf{y}\in\mathcal{R}^{n}\) be the observed \(n\) realizations of ternary and truncated variables, respectively. Let \(n_{0x}=\sum_{i=0}^{n}I(x_{i}=0)\), \(\pi_{0x}=\frac{n_{0x}}{n}\), \(n_{1x}=\sum_{i=0}^{n}I(x_{i}=1)\), \(\pi_{1x}=\frac{n_{1x}}{n}\), \(n_{2x}=\sum_{i=0}^{n}I(x_{i}=2)\), \(\pi_{2x}=\frac{n_{2x}}{n}\), \(n_{0y}=\sum_{i=0}^{n}I(y_{i}=0)\), \(\pi_{0y}=\frac{n_{0y}}{n}\), \(n_{0x0y}=\sum_{i=0}^{n}I(x_{i}=0 \;\& \; y_{i}=0)\), \(n_{1x0y}=\sum_{i=0}^{n}I(x_{i}=1 \;\& \; y_{i}=0)\) and \(n_{2x0y}=\sum_{i=0}^{n}I(x_{i}=2 \;\& \; y_{i}=0)\) then \[\begin{align} |\tau(\mathbf{x}, \mathbf{y})|\leq & \frac{\begin{pmatrix}n \\ 2\end{pmatrix}-\begin{pmatrix}n_{0x} \\ 2\end{pmatrix}-\begin{pmatrix}n_{1x} \\ 2\end{pmatrix}-\begin{pmatrix} n_{2x} \\ 2 \end{pmatrix}-\begin{pmatrix}n_{0y} \\ 2\end{pmatrix}+\begin{pmatrix}n_{0x0y} \\ 2 \end{pmatrix}+\begin{pmatrix}n_{1x0y} \\ 2\end{pmatrix}+\begin{pmatrix}n_{2x0y} \\ 2\end{pmatrix}}{\begin{pmatrix}n \\ 2\end{pmatrix}} \nonumber \end{align}\] Since \(n_{0x0y}\leq\min(n_{0x},n_{0y})\), \(n_{1x0y}\leq\min(n_{1x},n_{0y})\) and \(n_{2x0y}\leq\min(n_{2x},n_{0y})\) we obtain \[\begin{align} |\tau(\mathbf{x}, \mathbf{y})|\leq & \frac{\begin{pmatrix}n \\ 2\end{pmatrix}-\begin{pmatrix}n_{0x} \\ 2\end{pmatrix}-\begin{pmatrix}n_{1x} \\ 2\end{pmatrix}-\begin{pmatrix} n_{2x} \\ 2 \end{pmatrix}-\begin{pmatrix}n_{0y} \\ 2\end{pmatrix}}{\begin{pmatrix}n \\ 2\end{pmatrix}} \nonumber\\ & + \frac{\begin{pmatrix}\min(n_{0x},n_{0y}) \\ 2 \end{pmatrix}+\begin{pmatrix}\min(n_{1x},n_{0y}) \\ 2\end{pmatrix}+\begin{pmatrix}\min(n_{2x},n_{0y}) \\ 2\end{pmatrix}}{\begin{pmatrix}n \\ 2\end{pmatrix}} \nonumber\\ \leq & \frac{\begin{pmatrix}n \\ 2\end{pmatrix}-\begin{pmatrix}\max(n_{0x},n_{1x},n_{2x},n_{0y}) \\ 2\end{pmatrix}}{\begin{pmatrix}n \\ 2\end{pmatrix}} \nonumber\\ \leq & 1-\frac{\max(n_{0x},n_{1x},n_{2x},n_{0y})(\max(n_{0x},n_{1x},n_{2x},n_{0y})-1)}{n(n-1)} \nonumber\\ \approx & 1-(\frac{\max(n_{0x},n_{1x},n_{2x},n_{0y})}{n})^{2} \nonumber\\ =& 1-\{\max(\pi_{0x},\pi_{1x},\pi_{2x},\pi_{0y})\}^{2} \nonumber\\ =& 1-\{\max(\pi_{0x},(1-\pi_{0x}-\pi_{2x}),\pi_{2x},\pi_{0y})\}^{2} \end{align}\]
It is easy to get the approximate bound for truncated/ternary case by switching \(\mathbf{x}\) and \(\mathbf{y}\).